Ball and stick 4: Parallel vs serial mode
This is the final part of the series where we build a multicompartment cell and evolve it into a network of cells running on a parallel machine (which is basically all computers made within the last decade). On this page, we translate the classes we have previously constructed so that they operate in either a parallel or serial mode.
Note: If you followed the Quick Start instructions on the NEURON website, you should have everything you need to run parallel simulations. If not, if you do not already have an MPI installation, go to that link and follow the instructions in “Step 4: Install MPI to enable parallel simulations.” If you compiled NEURON yourself instead of using an installer (this is almost-never necessary), this part of the tutorial requires you to have used the --with-paranrn flag at configure time.
Parallel communication in NEURON
Parallel communication takes place via logical events in network connection objects known as NetCon. NetCon sources are threshold detectors. They monitor some variable, say the membrane potential of a cell, and when the variable reaches some threshold, it triggers an event sent to the targets. Targets are frequently synapses on other cells. When they receive the event, they activate.
In a parallel context across several machines, communication between hosts can be computationally inefficient when the frequency of events is high and when the message being sent is large. NEURON uses an efficient priority queueing mechanism to deliver events to targets after the delay specified by the NetCon. The message passed is succinct. It is an integer, the unique global identifier (gid) of the source. The following two figures illustrate these ideas and come from Hines M.L. and Carnevale N.T, Translating network models to parallel hardware in NEURON, Journal of Neuroscience Methods 169 (2008) 425–455. Users should also consult the ParallelContext reference.
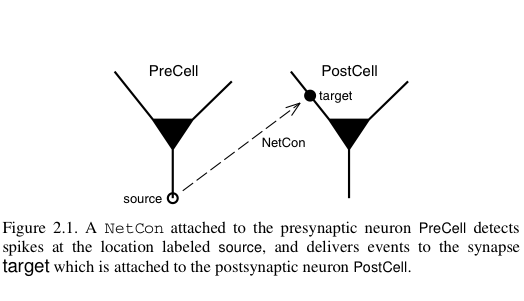
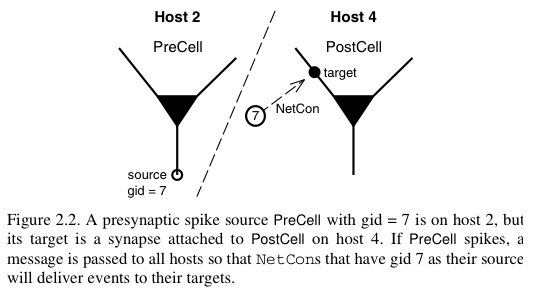
The main step involved in making a parallel implementation is to assign the global identifiers across the various hosts. Care should also be taken to assign cells to the various hosts such that the system is load balanced. For example, in a network with computationally complex and simple cells, several simple cells may be assigned to a host while few complex cells may be assigned to another host.
Test MPI and Parallel NEURON
Once again, parallel NEURON requires MPI support. If this is your first time using it in a while, you should test your computer setup.
Create a file called testmpi.py:
from neuron import h
h.nrnmpi_init() # initialize MPI
pc = h.ParallelContext()
print('I am {} of {}'.format(pc.id(), pc.nhost()))
h.quit() # necessary to avoid a warning message on parallel exit on some systems
Run the script in parallel via e.g. mpiexec -n 4 python testmpi.py from the command line in a terminal. You should see output resembling:
numprocs=4
I am 1 of 4
I am 2 of 4
I am 3 of 4
I am 0 of 4
These could appear in any order since in theory they are running simultaneously but must print out in some order. If instead you see four processes claiming to be 0 of 1, then your copy of NEURON was not compiled with support for parallel simulation. Reconfigure with the --with-paranrn flag, recompile, and try again. If you get an error saying that mpiexec is an unknown command, then MPI is either not installed or not on your PATH; correct your MPI setup and try again.
Back to the example
Begin by downloading ballandstick.py into your working directory. This is equivalent to the classes we created in the previous part of the tutorial.
We will construct a ring.py based on the previous Ring class. Changes are indicated with ###:
from neuron import h
from ballandstick import BallAndStick
### MPI must be initialized before we create a ParallelContext object
h.nrnmpi_init()
pc = h.ParallelContext()
class Ring:
"""A network of *N* ball-and-stick cells where cell n makes an
excitatory synapse onto cell n + 1 and the last, Nth cell in the
network projects to the first cell.
"""
def __init__(self, N=5, stim_w=0.04, stim_t=9, stim_delay=1, syn_w=0.01, syn_delay=5, r=50):
"""
:param N: Number of cells.
:param stim_w: Weight of the stimulus
:param stim_t: time of the stimulus (in ms)
:param stim_delay: delay of the stimulus (in ms)
:param syn_w: Synaptic weight
:param syn_delay: Delay of the synapse
:param r: radius of the network
"""
self._N = N
self.set_gids() ### assign gids to processors
self._syn_w = syn_w
self._syn_delay = syn_delay
self._create_cells(r) ### changed to use self._N instead of passing in N
self._connect_cells()
### the 0th cell only exists on one process... that's the only one that gets a netstim
if pc.gid_exists(0):
self._netstim = h.NetStim()
self._netstim.number = 1
self._netstim.start = stim_t
self._nc = h.NetCon(self._netstim, pc.gid2cell(0).syn) ### grab cell with gid==0 wherever it exists
self._nc.delay = stim_delay
self._nc.weight[0] = stim_w
def set_gids(self):
"""Set the gidlist on this host."""
#### Round-robin counting.
#### Each host has an id from 0 to pc.nhost() - 1.
self.gidlist = list(range(pc.id(), self._N, pc.nhost()))
for gid in self.gidlist:
pc.set_gid2node(gid, pc.id())
def _create_cells(self, r):
self.cells = []
for i in self.gidlist: ### only create the cells that exist on this host
theta = i * 2 * h.PI / self._N
self.cells.append(BallAndStick(i, h.cos(theta) * r, h.sin(theta) * r, 0, theta))
### associate the cell with this host and gid
for cell in self.cells:
pc.cell(cell._gid, cell._spike_detector)
def _connect_cells(self):
### this method is different because we now must use ids instead of objects
for target in self.cells:
source_gid = (target._gid - 1 + self._N) % self._N
nc = pc.gid_connect(source_gid, target.syn)
nc.weight[0] = self._syn_w
nc.delay = self._syn_delay
target._ncs.append(nc)
The call to h.nrnmpi_init() must happen before any use of the ParallelContext class – which forms a key part of any NEURON parallel simulation.
The only conceptually new method here is the set_gids method where each process specifies which cells it will simulate. Here we use what is known as a round-robin approach, where the pc.id()th process starts at pc.id() and skips by however many processes are running (pc.nhost).
In _create_cells, we now associate the cell gid with the NetCon _spike_detector. This allows the _connect_cells to make connections based on gids instead of objects, using pc.gid_connect.
Test the model
Here’s a basic test, test_ring1.py that loads the Ring class and plots cell 0’s membrane potential timeseries:
from neuron import h
from neuron.units import ms, mV
import matplotlib.pyplot as plt
from ring import Ring
cell_to_plot = 0
ring = Ring()
pc = h.ParallelContext()
pc.set_maxstep(10 * ms)
t = h.Vector().record(h._ref_t)
h.finitialize(-65 * mV)
pc.psolve(100 * ms)
if pc.gid_exists(cell_to_plot):
plt.figure()
plt.plot(t, pc.gid2cell(cell_to_plot).soma_v)
plt.show()
pc.barrier()
pc.done()
h.quit()
The code above should look very familiar.
The conceptually new pieces are:
pc.set_maxstep(10 * ms) – sets an upper bound on how far MPI can simulate without communicating, here a simulated 10 ms. This must be called before attempting a parallel simulation.
pc.psolve(100 * ms) – a parallel version of h.continuerun, but does not support updating NEURON graphics during the simulation.
pc.gid_exists – only the process that owns the specified cell should make the plot.
pc.gid2cell – lookup a cell by gid
pc.barrier() – wait until all processes reach this point; used to make sure processes don’t shut down before the graph is closed
Start by testing this without using MPI:
python test_ring1.py
You should see:
And once you are satisfied that works, you can try MPI, e.g.
mpiexec -n 2 python test_ring1.py
Gathering spikes
Our above test runs the simulation successfully, but in the end, no single process knows when all the spikes occurred. There are a number of ways to deal with this: one solution is to have each process write its data to a file. Instead, we will use pc.py_alltoall to send all the data to node 0, at which point node 0 can plot the raster, save data, or whatever.
Store this in test_ring2.py:
from neuron import h
from neuron.units import ms, mV
import matplotlib.pyplot as plt
from ring import Ring
ring = Ring()
pc = h.ParallelContext()
pc.set_maxstep(10 * ms)
t = h.Vector().record(h._ref_t)
h.finitialize(-65 * mV)
pc.psolve(100 * ms)
# send all spike time data to node 0
local_data = {cell._gid: list(cell.spike_times) for cell in ring.cells}
all_data = pc.py_alltoall([local_data] + [None] * (pc.nhost() - 1))
if pc.id() == 0:
# combine the data from the various processes
data = {}
for process_data in all_data:
data.update(process_data)
# plot it
plt.figure()
for i, spike_times in data.items():
plt.vlines(spike_times, i + 0.5, i + 1.5)
plt.show()
pc.barrier()
pc.done()
h.quit()
Running this via e.g. mpiexec -n 2 python test_ring2.py displays the familiar raster plot. If you are wondering why node 0 was the one chosen to make the plot, it is because that is the only node that is guaranteed to exist (nothing else exists if there is only one process being used for the simulation).
You should see:
[ ]: